Top 10 Publications
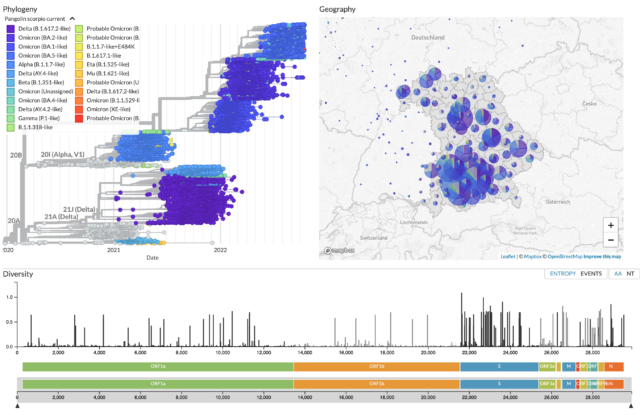
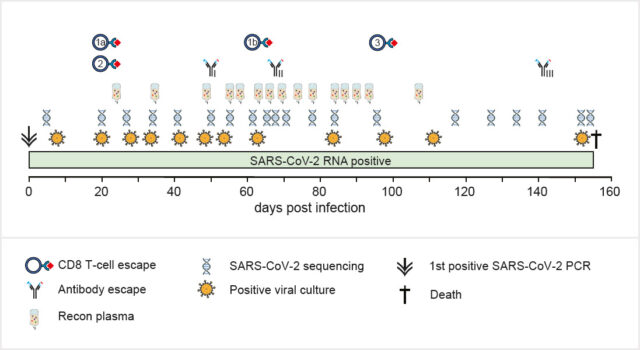
Khatamzas E, Antwerpen M H, Rehn A, Graf A, Hellmuth J C, Hollaus A, Mohr A W, Gaitzsch E, Weiglein T, Georgi E, Scherer C, Stecher S S, Gruetzner S, Blum H, Krebs S, Reischer A, Leutbecher A, Subklewe M, Dick A, Zange S, Girl P, Muller K, Weigert O, Hopfner K P, Stemmler H J, von Bergwelt-Baildon M, Keppler O T, Wolfel R, Muenchhoff M*, Moosmann A*. Accumulation of mutations in antibody and CD8 T cell epitopes in a B cell depleted lymphoma patient with chronic SARS-CoV-2 infection. Nature Communications 13:5586 (2022). https://doi.org/10.1038/s41467-022-32772-5
Muenchhoff M, Graf A, Krebs S, Quartucci C, Hasmann S, Hellmuth J C, Scherer C, Osterman A, Boehm S, Mandel C, Becker-Pennrich A S, Zoller M, Stubbe H C, Munker S, Munker D, Milger K, Gapp M, Schneider S, Ruhle A, Jocham L, Nicolai L, Pekayvaz K, Weinberger T, Mairhofer H, Khatamzas E, Hofmann K, Spaeth P M, Bender S, Kaab S, Zwissler B, Mayerle J, Behr J, von Bergwelt-Baildon M, Reincke M, Grabein B, Hinske C L, Blum H, Keppler O T. Genomic epidemiology reveals multiple introductions of SARS-CoV-2 followed by community and nosocomial spread, Germany, February to May 2020. EuroSurveillance 26 (2021). https://doi.org/10.2807/1560-7917.ES.2021.26.43.2002066
Muenchhoff M, Mairhofer H, Nitschko H, Grzimek-Koschewa N, Hoffmann D, Berger A, Rabenau H, Widera M, Ackermann N, Konrad R, Zange S, Graf A, Krebs S, Blum H, Sing A, Liebl B, Wolfel R, Ciesek S, Drosten C, Protzer U, Boehm S, Keppler O T. Multicentre comparison of quantitative PCR-based assays to detect SARS-CoV-2, Germany, March 2020. EuroSurveillance 25 (2020). https://doi.org/10.2807/1560-7917.ES.2020.25.24.2001057
Muenchhoff M, Chung A W, Roider J, Dugast A S, Richardson S, Kloverpris H, Leslie A, Ndung'u T, Moore P, Alter G, Goulder P J R. Distinct Immunoglobulin Fc Glycosylation Patterns Are Associated with Disease Nonprogression and Broadly Neutralizing Antibody Responses in Children with HIV Infection. mSphere 5 (2020). https://doi.org/10.1128/mSphere.00880-20
Muenchhoff M, Adland E, Roider J, Kloverpris H, Leslie A, Boehm S, Keppler O T, Ndung'u T, Goulder P J R. Differential Pathogen-Specific Immune Reconstitution in Antiretroviral Therapy-Treated Human Immunodeficiency Virus-Infected Children. Journal of Infectious Diseases 219:1407-1417 (2019). https://doi.org/10.1093/infdis/jiy668
Muenchhoff M, Healy M, Singh R, Roider J, Groll A, Kindra C, Sibaya T, Moonsamy A, McGregor C, Phan M Q, Palma A, Kloverpris H, Leslie A, Bobat R, LaRussa P, Ndung'u T, Goulder P, Sobieszczyk M E, Archary M. Malnutrition in HIV-Infected Children Is an Indicator of Severe Disease with an Impaired Response to Antiretroviral Therapy. AIDS Research and Human Retroviruses 34:46-55 (2018). https://doi.org/10.1089/AID.2016.0261
Muenchhoff M, Adland E, Karimanzira O, Crowther C, Pace M, Csala A, Leitman E, Moonsamy A, McGregor C, Hurst J, Groll A, Mori M, Sinmyee S, Thobakgale C, Tudor-Williams G, Prendergast A J, Kloverpris H, Roider J, Leslie A, Shingadia D, Brits T, Daniels S, Frater J, Willberg C B, Walker B D, Ndung'u T, Jooste P, Moore P L, Morris L, Goulder P. Nonprogressing HIV-infected children share fundamental immunological features of nonpathogenic SIV infection. Science Translational Medicine 8:358ra125 (2016). https://doi.org/10.1126/scitranslmed.aag1048
Payne R*, Muenchhoff M*, Mann J, Roberts H E, Matthews P, Adland E, Hempenstall A, Huang K H, Brockman M, Brumme Z, Sinclair M, Miura T, Frater J, Essex M, Shapiro R, Walker B D, Ndung'u T, McLean A R, Carlson J M, Goulder P J. Impact of HLA-driven HIV adaptation on virulence in populations of high HIV seroprevalence. Proceedings of the National Academy of Science (PNAS) 111:E5393-400 (2014). https://doi.org/10.1073/pnas.1413339111
Muenchhoff M, Prendergast A J, Goulder P J. Immunity to HIV in Early Life. Frontiers in Immunology 5:391 (2014). https://doi.org/10.3389/fimmu.2014.00391
Muenchhoff M, Goulder P J. Sex differences in pediatric infectious diseases. Journal of Infectious Diseases 209 Suppl 3:S120-6 (2014). https://doi.org/10.1093/infdis/jiu232