Research Group Prof. Dr. Bärbel Stecher
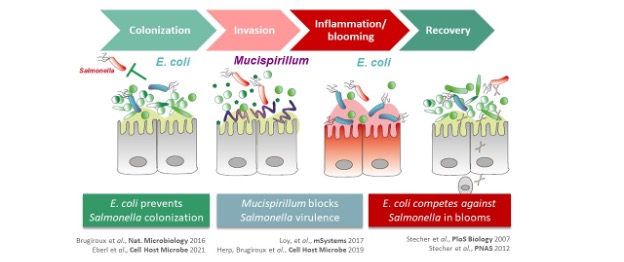
The human gastrointestinal tract is an ecosystem of unsurpassed diversity. Microbial communities termed microbiota, including bacteria, archaea, eukaryotes and viruses colonize intestine. In particular, the bacterial members engage in a physiological network of cooperation and competition via their metabolic products, which has an enormous influence on human health. In otherwise healthy human individuals, the microbiota is mostly able to block colonization of enteric pathogens in a process termed colonization resistance. However, antibiotics, immunosuppression and inflammation can disrupt colonization resistance and introduce microbiota imbalances with increased susceptibility to pathogen infections. A disturbed microbiota is characterized by depletion of the obligate anaerobic species and a relative enrichment in facultative anaerobic bacteria, in particular members of the Enterobacteriaceae, leading to the term of “Enterobacterial blooming”. The mechanisms driving inflammation-induced blooms include generation of oxygen and alternative electron acceptors (e. g. NO3-), production of antimicrobials, iron depletion and intraluminal phagocyte-killing. Inflammation-induced Enterobacterial blooms are a potential threat to human health by increasing the risk of systemic infections, promoting horizontal spread of fitness- and virulence factors, as well as antibiotic resistances among potentially pathogenic species.
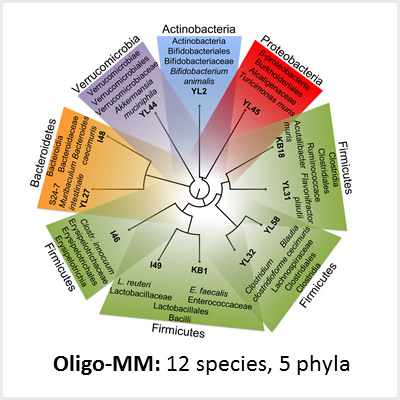
Due to the enormous complexity of the intestinal microbiota and the lack of appropriate animal models, the mechanisms governing colonization resistance, i. e. interaction between microbiota, host immune system and pathogens, remain largely unclear. We use synthetic microbial communities which we can study in vitro and in gnotobiotic mice. With a focus on infections with Gram-negative enteric pathogens including Salmonella enterica and pathogenic Escherichia coli, we aim for the following:
- Understanding interaction of commensal bacteria with each other and with enteric pathogens
- Elucidating how phages, inflammatory immune responses and antibiotics affect the microbiota and pathogens, their interaction and gene expression.
- Analysing how bacterial evolution and horizontal gene transfer in the intestinal ecosystem is shaped by microbial interactions, environmental factors and interaction with the host´s mucosa