Top 10 Publications
Ailloud F, Estibariz I, Pfaffinger G, Suerbaum S (2022) The Helicobacter pylori UvrC nuclease is essential for chromosomal micro-imports after natural transformation. mBio 13:e0181122.
https://doi.org/10.1128./mbio.01811-22.
Suerbaum S, Coombs N, Patel L, Pscheniza D, Rox K, Falk C, Gruber AD, Kershaw O, Chhatwal P, Brönstrup M, Bilitewski U, Josenhans C (2022) Identification of antimotilins, novel inhibitors of Helicobacter pylori flagellar motility that inhibit stomach colonization in a mouse model. mBio 13:e0375521.
https://doi.org/10.1128/mbio.03755-21.
Ailloud F, Estibariz I, Pfaffinger G, Suerbaum S (2022) The Helicobacter pylori UvrC nuclease is essential for chromosomal micro-imports after natural transformation. mBio 13:e0181122. https://doi.org/10.1128./mbio.01811-22.
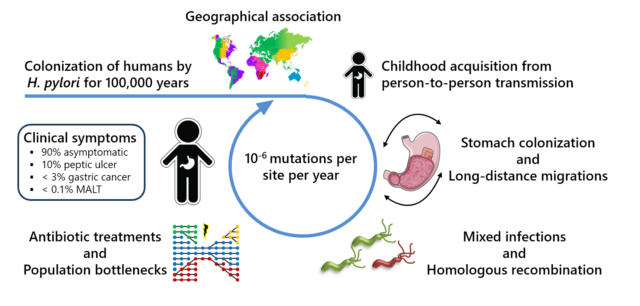
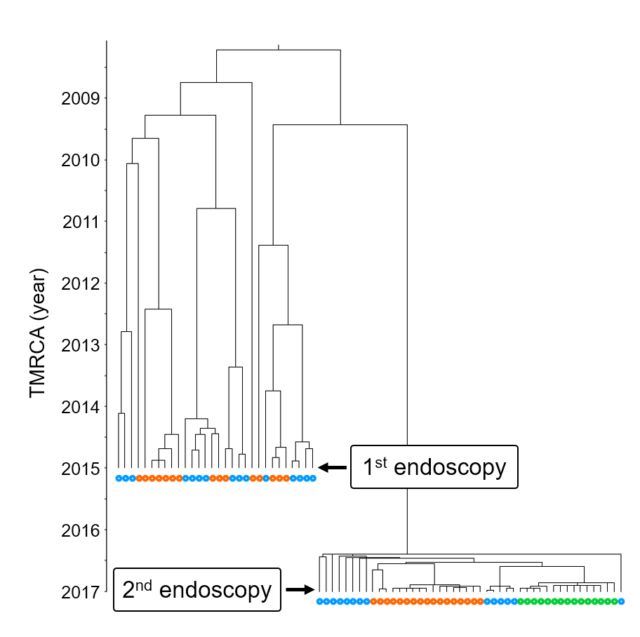
Ailloud, F., X. Didelot, S. Woltemate, G. Pfaffinger, J. Overmann, R. C. Bader, C. Schulz, P. Malfertheiner, S. Suerbaum. 2019. Within-host evolution of Helicobacter pylori: niche-specific adaptation, migrations between gastric locations, and selective sweeps. Nat. Commun. 10:2273.
https://doi.org/10.1038/s41467-019-10050-1.
Nell S, Estibariz I, Krebes J, Bunk B, Graham DY, Overmann J, Song Y, Spröer C, Yang I, Wex T, Korlach J, Malfertheiner P, Suerbaum S (2018) Genome and methylome variation in Helicobacter pylori with a cag pathogenicity island during early stages of human infection. Gastroenterology 154:612.
https://doi.org/10.1053/j.gastro.2017.10.014.
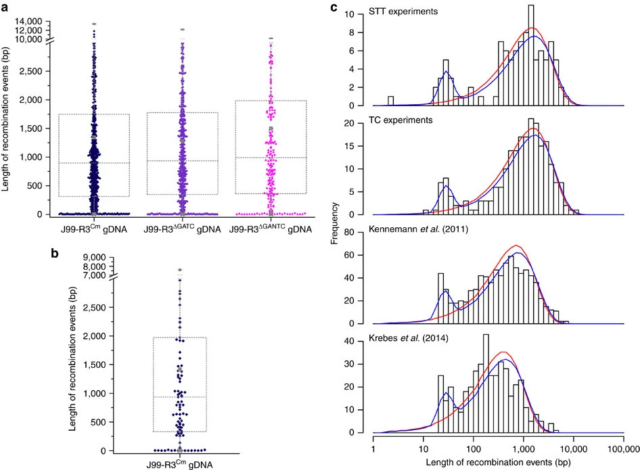
Bubendorfer S, Krebes J, Yang I, Hage E, Schulz TF, Bahlawane C, Didelot X, Suerbaum S (2016) Genome-wide analysis of chromosomal import patterns after natural transformation of Helicobacter pylori. Nat Commun 7:11995.
https://doi.org/10.1038/ncomms11995.
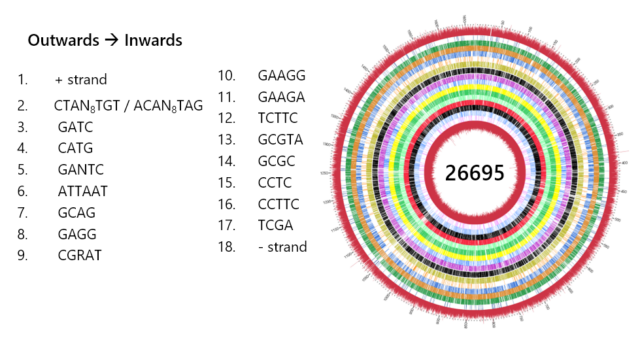
Krebes J, Morgan RD, Bunk B, Spröer C, Luong K, Parusel R, Anton BP, König C, Josenhans C, Overmann J, Roberts RJ, Korlach J, Suerbaum S (2014) The complex methylome of the human gastric pathogen Helicobacter pylori. Nucl Acids Res 42:2415.
https://doi.org/10.1093/nar/gkt1201.
Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, Reinhardt R, Correa P, Meyer TF, Josenhans C, Falush D, & Suerbaum S (2011) Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci USA 108:5033.
https://doi.org/10.1073/pnas.1018444108.
Linz B, Balloux F, Moodley Y, Manica A, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, Yamaoka Y, Graham DY, Perez-Trallero E, Wadstrom T, Suerbaum S, Achtman M (2007) An African origin for the intimate association between humans and Helicobacter pylori. Nature 445:915.
https://doi.org/10.1038/nature05562.
Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, Yamaoka Y, Mégraud F, Otto K, Reichard U, Katzowitsch E, Wang X, Achtman M, Suerbaum S (2003) Traces of human migrations in Helicobacter pylori populations. Science 299: 1582.
https://doi.org/10.1126/science.1080857.
Suerbaum S, Michetti P (2002) Helicobacter pylori infection. N Engl J Med 347:1175 https://doi.org/10.1056/NEJMra020542 (Review).