Phages are a diverse group of viruses that can infect bacterial cells. As any other virus upon infection and entry into the cell, phages turn the bacterium into a factory designed to produce viral particles which ultimately lyse the bacterium to enter the environment and start a new round of infection (lytic infection). Temperate phages are a specific group of phages characterised by their ability to integrate their genetic material into the bacterial genome (lysogenic infection). In this state they can reside as prophages for many generations along with the host. However, prophages are molecular time bombs that can switch from this dormant state to the lytic state. This switch can happen either spontaneously or in response to environmental stress, such as DNA damage.
Prophages can influence bacterial ecology and evolution in various ways that often extend beyond the bacterium and translate via cascading effects to higher organisms and even entire ecosystems (Wendling 2023). These include beneficial impacts, such as accessory genes encoded on prophages which can for instance alter bacterial pathogenicity, but also negative impacts, such as the risk of lysis. In our group we study how these prophage-mediated impacts affect their ecological and evolutionary interactions with bacteria and how this influences human health and disease.
The role of prophages in the spread and maintenance of antibiotic resistance genes
Antibiotic resistance genes (ARGs) are often encoded on mobile genetic elements (MGEs), including plasmids and phages. The spread of antibiotic resistance through MGEs poses a significant threat to treating infections, making it crucial to understand how MGEs transfer and interact with each other in natural environments. Most studies focus on MGEs in isolation without considering their interactions. In a recent study, we combined in vitro experiments and mathematical modeling to reveal the influence of prophages on conjugation. We found that prophages can significantly restrict the spread of conjugative plasmids, with this effect varying based on environmental conditions and bacterial genetics (Igler et al. 2022). Simulating a range of infection parameters showed that high conjugation rates could enable plasmids to overcome the negative impact of prophages.
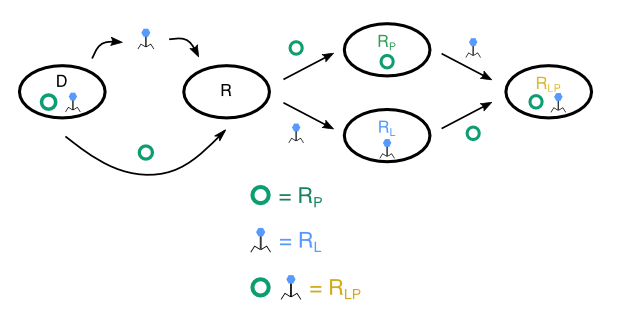
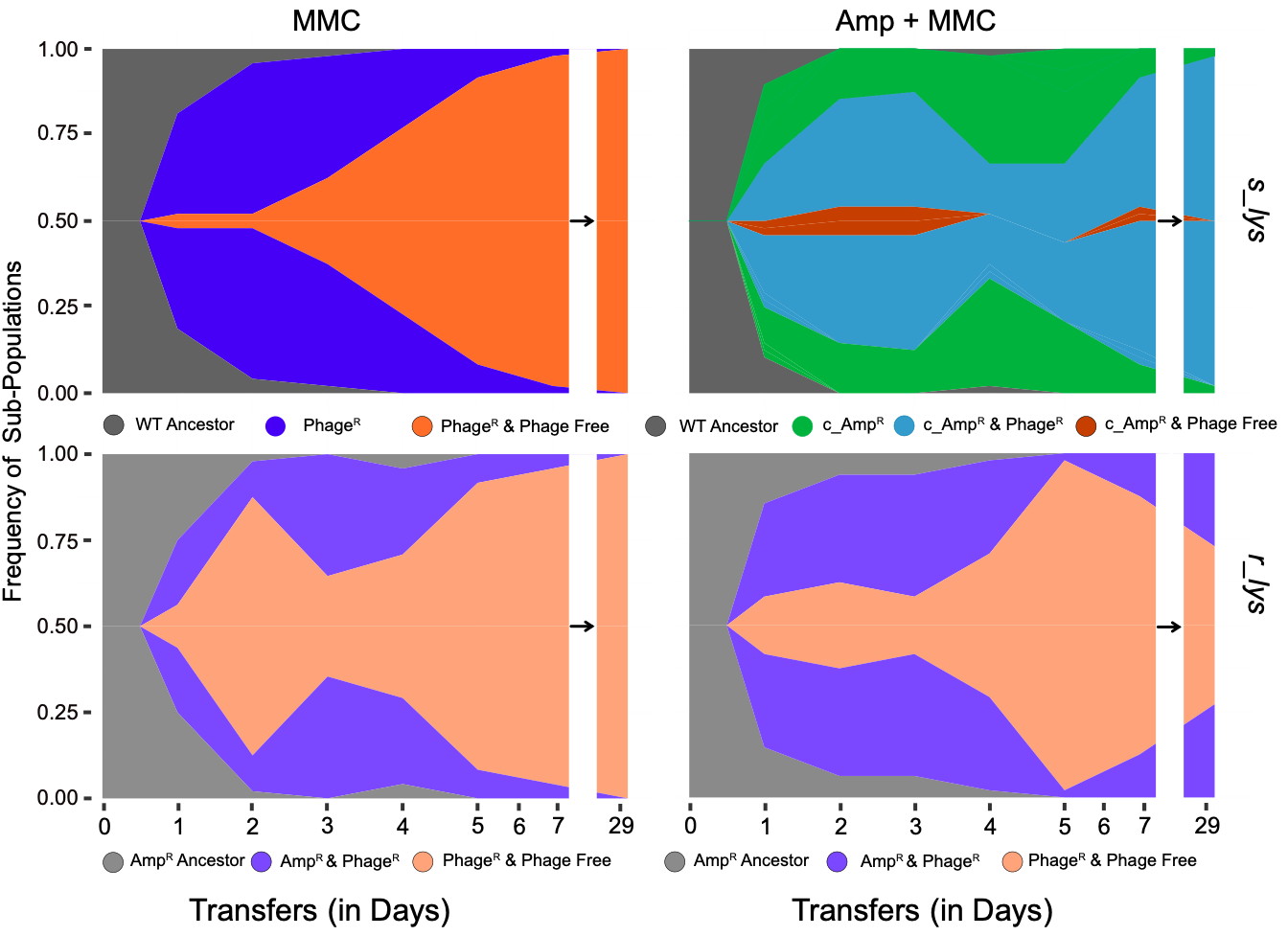
In addition to their spread, the maintenance of MGEs carrying ARGs is of great interest. While prophages encoding ARGs can be highly beneficial for bacteria in the presence of antibiotics (Wendling et al. 2020), the ability of prophages to switch back to the lytic cycle puts bacteria at constant risk of lysis and death. Our group studies how these contrasting fitness impacts of prophages affect their ecological and evolutionary interactions with bacteria. Using experimental evolution, we found that prophages quickly disappear from a bacterial population in environments that enhance the switch from the lysogenic to the lytic cycle. In contrast, prophages carrying beneficial genes can persist despite frequent induction (Bailey et al., 2024).
Bailey, Z.M., Igler, C., Wendling, C.C*. (2024) „Prophage maintenance is determined by environment-dependent selective sweeps rather than mutational availability” Current Biology https://doi.org/10.1016/j.cub.2024.03.025
Igler, C., Schwyter, L., Gehrig, D., Wendling, C.C.* (2022) „Conjugative plasmid transfer is limited by prophages but can be overcome by high conjugation rates” Phil Trans B. (377)1842
https://doi.org/10.1098/rstb.2020.0470
Wendling, C.C.*, Refardt, D., Hall, A.R. (2020) “Fitness benefits to bacteria of carrying prophages and prophage-encoded antibiotic-resistance genes peak in different environments” Evolution 75(2) https://doi.org/10.1111/evo.14153
Funding: SNF Ambizione 179743
Collaborators: Claudia Igler, Alex Hall, Dominik Refardt
Drivers and Constraint of Prophages in Natural Communities:
Phage lambda has a spontaneous excision rate of 106 per cell division under non-stressful conditions, which can increase by orders of magnitude under stressful conditions. Thus, it is predicted that functional prophages will not persist in the bacterial chromosome for an extended time period. In a recent study, we could confirm these predictions by demonstrating that, driven by environment-specific sequences of selective sweeps, prophages can rapidly disappear from a lysogen population, if prophage-associated costs outweigh their benefits (Bailey at al. 2024). While our study highlights the complexity of selection pressures that act on functional prophages in vitro, we know little about the long-term maintenance of functional prophages in natural populations. This project explores the persistence and evolution of prophages in natural bacterial communities. To do so, we are conducting a prospective cohort study during which we are repeatedly sampling resident E. coli from human gut microbiomes. By combining the cohort study with in vitro gut mesocosms experiments and advanced bioinformatic analyses we address fundamental questions in prophage ecology and evolution: are prophages persistent or transient? Which ecological and evolutionary factors promote or constrain prophage persistence in natural communities over time?
Bailey, Z.M., Igler, C., Wendling, C.C*. (2024) „Prophage maintenance is determined by environment-dependent selective sweeps rather than mutational availability” Current Biology https://doi.org/10.1016/j.cub.2024.03.025
https://spp2330.de/project-wendling/
Funding: SNF Ambizione 179743
Collaborators: Claudia Igler, Anne Kupczok
Understanding phage resistance evolution to advance phage therapy:
Amid the antibiotic resistance crisis, the century-old practice of phage therapy, which uses bacterial viruses to treat bacterial infections, has gained renewed attention. Despite its advantages over antibiotics and extensive research, it has yet to gain momentum due to remaining limitations. These include for instance the evolution of bacterial resistance to therapeutic phages. This is a growing concern which prompts the exploration of various strategies such as directing phage-resistance evolution, which can result in beneficial fitness trade-offs, such as reduced virulence or antibiotic susceptibility. Using experimental evolution, we found that phage resistance correlates negatively with bacterial virulence (Wendling et al. 2022). That is, because many phages utilize the same molecular receptor for bacterial cell entry as the pathogen itself uses for host attachment or motility, which may include structures like flagella or pili (Wendling et al. 2022, Goehlich et al. 2023). Such a shared molecular structure creates a unique scenario where the evolution of phage resistance, i.e., modification of the pilus or flagellum, could impose pleiotropic costs in terms of reduced virulence (Wendling et al. 2017, Wendling et al, 2022, Goehlich et al. 2023).
Funding: DFG Future Ocean Excellence Cluster, DFG SPP1819
Collaborators: Olivia Roth, Heiko Liesegang, Mike Brockhurst
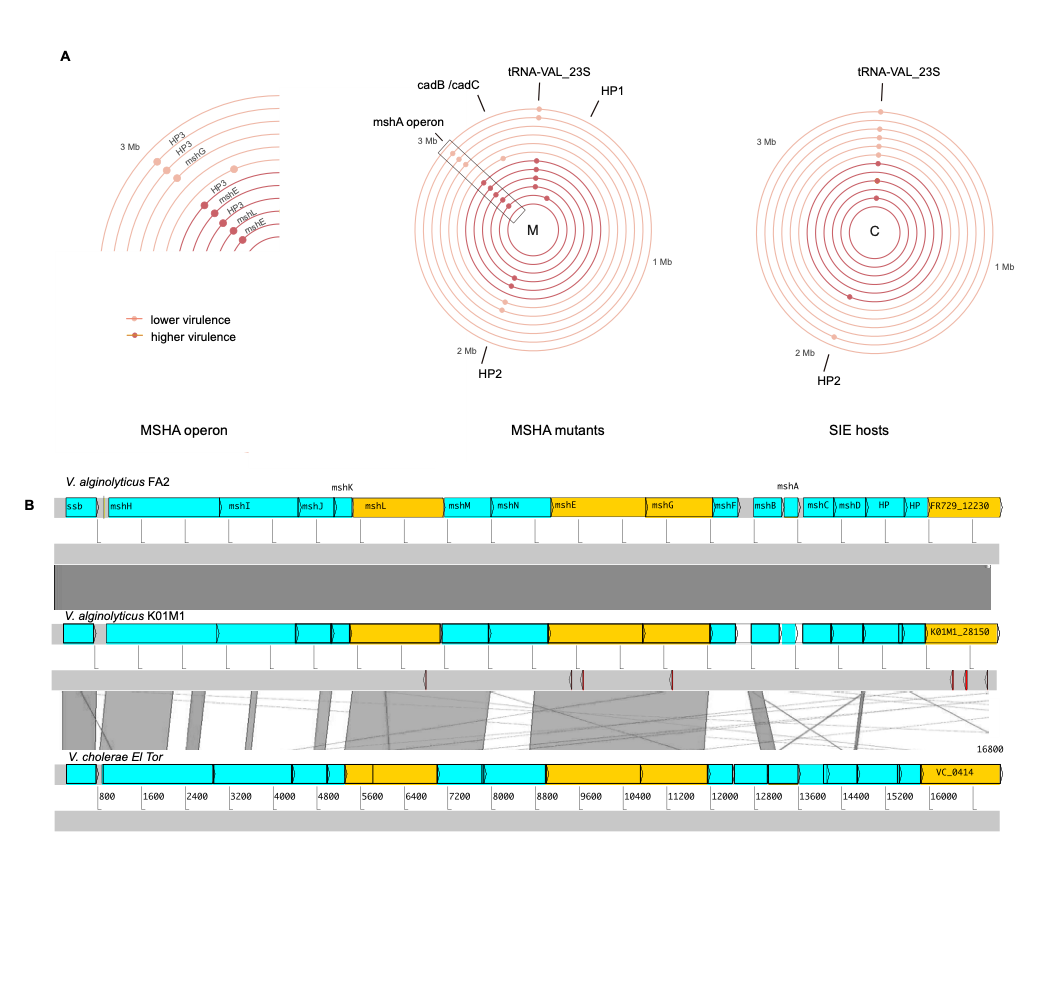
Wendling, C.C.*, Piecyk, A., Refardt, D., Chibani, C.M., Hertel, R., Liesegang, H. Bunk, B., Overmann, J., Roth, O. (2017): “Tripartite species interaction: Eukaryotic hosts suffer more from phage susceptible than from phage resistant bacteria”. BMC Evolutionary Biology
https://bmcevolbiol.biomedcentral.com/articles/10.1186/s12862-017-0930-2
Wendling, C.C.*, Lange, J., Liesegang, H., Sieber, M., Poehlein, A., Bunk, B., Rajkov, J., Goehlich, H., Roth, O., Brockhurst, M. (2022) „Higher phage virulence accelerates the evolution of host resistance” Proceeding of the Royal Society B (289)1984 https://doi.org/10.1098/rspb.2022.1070
Goehlich, H., Roth, O., Sieber, M., Chibani, C.M., Rajkov, J., Liesegang, H., Wendling, C.C.* (2023) „Sub-optimal environmental conditions prolong phage epidemics.” Molecular Ecology
https://doi.org/10.1111/mec.17050
Phage-Pro: Advancing Phage Therapy through synergistic strategies
Amid the antibiotic resistance crisis, the century-old practice of phage therapy, the use of bacterial viruses to treat bacterial infections, has gained renewed attention. Despite extensive research and clear-cut advantages over antibiotics, phage therapy has yet to gain momentum. This is primarily due to remaining limitations, e.g., the time-consuming process of identifying suitable phages and limited in-vivo efficacy.
In the EU-funded project Phage-Pro, we will explore a new technology to overcome the shortcomings of traditional phage therapy. Unlike the conventional use of lytic phages, we will capitalize on prophages – viruses that integrate into bacterial genomes capable of excision and replication. By integrating prophages into safe carrier strains, originating from probiotics, we plan to unlock three pivotal benefits, (1) rapid identification of suitable phages using advanced machine learning, (2) sustained in-vivo efficacy, and (3) the synergistic advantage of phage killing and pathogen competitive exclusion imposed by the probiotic.
Funding: ERC Starting Grant

https://www.lmu.de/en/newsroom/news-overview/news/six-new-erc-grants-at-lmu.html
https://www.helmholtz-munich.de/newsroom/interviews/erc-grants-2024